GRAPHICAL INSPECTION OF PET RESULTS IN SPM96 - A STEP BY STEP GUIDE
Index
General selection of contrast/s - to set up viewing results.
Maxima - finding specific peaks, to inspect statistically and graphically.
Plot - drawing various types of histogram.
Slices - creating transverse slices around a particular activation.
Sections - as slices, but in the three different plains.
Write - create files of your images.
Render - overlay your activations on a 3D brain.
Manipulating the output in the Graphics Window - changing the format and printing it.
Technical notes - notes on miscellaneous technical issues.
General selection of contrast/s
Within Unix move to the directory that your analysis resides.
Type SPM.
Choose "PET and SPECT" option.
There are various ways to view and present your analysis results. Most of these options can be reached via the "Results" button, in the top left hand window.
Press "Results."
When asked "which SPM" choose "SPM(Z)."
The SPMget dialog box opens, looking for the SPMt.mat file. Hopefully (if your desired analysis directory is showing) this will be the only file listed. If so, click on it to highlight it and press "Done." Otherwise navigate (as outlined in the standard analysis section) to the right directory and then select the required file.
When asked for "conjunction analysis" choose "no." (Yes would combine contrasts in your analysis together)
Then pick the contrast of the analysis that you are interested in.
When asked "Mask with other contrast/s," pick "no." (Yes would mean that voxels that didn't survive correction in one or more other contrast would be eliminated from presentation for this contrast).
You are then asked for the "height threshold." This is an uncorrected Z score threshold. If you type a number here greater than one, then spm understands this as a Z score, and eliminates from all further display any voxels with Z scores of less than this value. If you type a number that is less than 1, then spm will understand this an uncorrected p value, and eliminate from display any voxels with a Z score that is associated with a p value larger than (nearer 1) the p value you have typed (see Z scores and p values for more information on the relationship of Z scores and p values). Note that, for the voxel to be displayed at any threshold, it must have survived the thresholding applied during the original analysis. A very large number of voxels are thrown away during the analysis, either because they did not survive the gray matter thresholding, or because they did not have a sufficiently high F value at the F statistic thresholding.
The default output, as put in the spm.ps file, is p (uncorrected probability) = 0.01. (If you haven't already, you can view this output by moving to the analysis directory and typing "ghostview spm.ps" at the Unix prompt. See here for information about ghostview and reading postscript files). If your region of activation of particular interest was not present on this output file because the Z value was too low (i.e it wasn't significantly active, given the output default probability threshold), then you can correct that now by choosing a higher p value (p nearer 1) for this "height threshold" input. For instance you could choose p=0.05 or p=0.1. Remember, this can always be modified by re-running the above steps if it isn't quite the right threshold level when the images are created. On the other hand, if your activation on the default output in spm.ps is a very significant peak, but there is also an awful lot of other activation areas of lower significance, you might like to change the threshold in the opposite direction, graphically to remove the other unwanted areas and leave your interested areas intact. The current level (as the results default in front of you) of 0.001 will achieve this, although you might like to try an even more strict threshold, such as 0.0001. Since this is only modifying the graphical presentation of your results, the exact value you choose isn't too important, We recommend that, when you first view your results, you choose a liberal height threshold, such as p<0.01, to screen your data for effect, but that (in general) you use a more conservative threshold when presenting your results. For example you might consider a corrected height threshold of p<0.1 (see corrected height thresholding for how to do this).

When asked for "Use corrected extent threshold" press "no." (If you press "yes", then the threshold will be a p value (that you subsequently specify) corrected for extent. See corrected extent thresholding for more explanation).
The next question spm asks (if you chose No for corrected for extent), is for an extent threshold. This may seem to be the same, but is an uncorrected extent threshold. You can enter here a value greater than one, or less than one. If you enter a value greater than one, say X, then spm will simply remove all clusters that have less than X voxels. If you enter a number less than one, spm will assume this is a probability, and will work out how many voxels the clusters should be, based on this p value, using some rather complex maths. We recommend you do one of (in order of preference):
- Type 1 to turn off the extent thresholding, and see all your data, or;
- Type some number greater than one, to conceal very small clusters, of size (in voxels) of less than the number you typed.
The graphics window on the right hand side now produces the results output for your specifications. In the top there are three views of the brain. The views are (clockwise from top left) sagittal, coronal and transverse. These views are known as glass brains, since they present activations in the three views as if the brain were transparent (see MIPs below for details). To fix the location of an activation area that you are interested in, it is useful to trace an activation from one of the images horizontally or vertically along to the other image, and then to the third image, so that you can determine the three-dimensional location of this area. There are various ways of viewing these results and manipulating them graphically in this window:
Maxima (looking at significant activations in a given contrast):
There is a red arrow, that starts by default in the middle of the brain images. This can be moved to a particular activation in two ways. First of all it can be dragged by holding the left mouse button over it and moving the arrow to a particular place in the images. For example, you could use the red arrow on the sagittal image to move the arrow in X and Z, then use the arrow on the transverse view to move the arrow in Y.
Alternatively, co-ordinates can be entered into in the top left of the graphics window. The x co-ordinate moves the cursor left (negative) or right (positive) in the brain. The y co-ordinate moves the cursor in an anterior direction (positive) or posterior direction (negative) and the z co-ordinate moves the cursor in a dorsal direction (positive) or a ventral direction (negative). Once the cursor is in the rough area of the activation, press the "maxima" button on the left of the graphics window. The nearest significant activation peak will be chosen, and statistical information for that and other nearby peaks will appear at the bottom of the window (see the SPM statistics and random fields tutorials for information on the meaning of the various figure columns).
 Plot (creating graphs looking at various aspects of the data):
Plot (creating graphs looking at various aspects of the data):
Once a maximum has been chosen by the above method, select the button "plot" on the left-hand-side of the graphics window (on the right of the screen). Pressing "conditions" in the lower left-hand window will produce a histogram with each x-axis number being a condition, and the size of the bar (y-axis) being the blood flow for the peak chosen. For other options, choose "specify." Three further options are presented to you. "Cov" refers to a plot for one of your covariates (if you have any) for this brain region. "Scan" produces a plot giving the activity in that area for each scan for each subject. The last option ("specify") allows you to plot the data against a covariate that you provide in the SPM input window.
Slices (creating three transverse slices of the specified area of activation):
 Once a maximum has been chosen by the above method, select the button "slices" on the left-hand-side of the graphics window (on the right of the screen). The SPMget dialog box will open and you will be asked to "select an image for rendering." Click on the "previous directories…" wide button towards the top of this dialog box, and choose the lowest option, which should be the spm main directory. This will usually be /cbu/imagers/spm/spm96. Then click on the "canonical" directory. Choose "T1.img." If you can't find the canonical directory in the main spm directory at the bottom of the list, then try going to the /cbu/imagers/spm/spm96 directory. T1.img is an unsmoothed image of the individual brain considered most typical of the 305 brains used to define the MNI standard. Three slices will be produced at the bottom of the graphics window. The middle slice corresponds exactly to the z value (height) of your maximum, while the one on the left is one slice ventral to this and the one on the right is one slice dorsal to this. The red cross shows precisely where the peak of activation is.
Once a maximum has been chosen by the above method, select the button "slices" on the left-hand-side of the graphics window (on the right of the screen). The SPMget dialog box will open and you will be asked to "select an image for rendering." Click on the "previous directories…" wide button towards the top of this dialog box, and choose the lowest option, which should be the spm main directory. This will usually be /cbu/imagers/spm/spm96. Then click on the "canonical" directory. Choose "T1.img." If you can't find the canonical directory in the main spm directory at the bottom of the list, then try going to the /cbu/imagers/spm/spm96 directory. T1.img is an unsmoothed image of the individual brain considered most typical of the 305 brains used to define the MNI standard. Three slices will be produced at the bottom of the graphics window. The middle slice corresponds exactly to the z value (height) of your maximum, while the one on the left is one slice ventral to this and the one on the right is one slice dorsal to this. The red cross shows precisely where the peak of activation is.
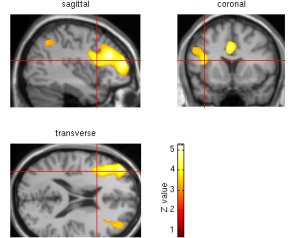 Sections (creating slices in the three different dimensions for the specified area of activation):
Sections (creating slices in the three different dimensions for the specified area of activation):
Once a maximum has been chosen by the above method, select the button "sections" on the left-hand-side of the graphics window (on the right of the screen). The SPMget dialog box will open and you will be asked to "select an image for rendering." As with slices, click on the "previous directories…" wide button towards the top of this dialog box, and choose the lowest option, which should be /cbu/imagers/spm/spm96 (but see above). Then click on the "canonical" directory. Choose "T1.img." At the bottom of the graphics window on the right, three images will be created, one transverse (as slices), one sagittal, and one coronal - centring on the co-ordinates of your specified maximum. The red cross, as with slices, shows the precise peak of activation of the maximum chosen.
Write (writing files for the results):
Choosing this option writes the results presented in the graphics window to a file in the directory you opened SPM from. This file can be read by SPM (such as in the display option on the upper left window), or Analyze. Specify the filename at the prompt in the lower left window. Also see " Print " below.
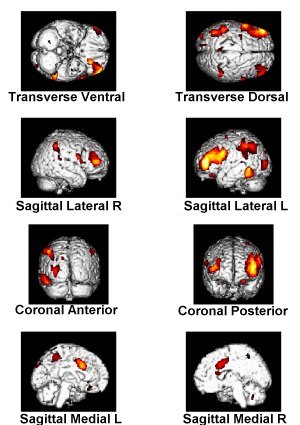 Render (overlays all activations for a contrast on the rendered three dimensional brain):
Render (overlays all activations for a contrast on the rendered three dimensional brain):
In the bottom section of the top left-hand window on the screen, click on the "render" button. Choose "SPM(Z)." The SPMget dialog box appears, asking for the SPMt.mat file. If this isn't already shown, navigate as instructed in the standard SPM analysis page until the directory of your analysis of interest is shown (the best way to do this is probably via the "previous directories" button towards the top of this dialog box). Select the SPMt.mat file, and press "done." Repeat all the steps above concerning the specifications of the graphical presentation of your results for a given contrast (or conjunction of contrasts). Once this is completed, on the graphics window on the right, eight images of brains will be presented. The legends to the images on the left illustrate what each of the eight images present. For each of these images, the areas of activation for this contrast (or conjunction of contrasts) will be overlaid on this standard 3D rendered brain.
Manipulating the output in the Graphics Window:
Print:
Prints the exact appearance of the current graphics window to another page of the spm.ps file, where it can be highlighted and selectively printed (see the SPM tips page for details). Note that pressing this "print" button doesn't print the graphical output to any real printer, but to the spm.ps file, by default.
Clear:
Clears the graphical window of all images and objects. There is very little point of ever pressing this button, since any new graphical output automatically clears the graphical window.
Colormap:
Pressing "colormap" at the top of the graphics window will produce a drop-down menu of three possibilities:
Gray - everything in black and white
Hot - all brains in colours (a little confusing)
Split - the brain in black and white, but the activations in colour (the default).
Effects:
Pressing "effects" at the top of the graphics window will produce a drop-down menu of three possibilities:
Invert - inverts the colours of the colormap option. So if the colormap is at default, the brains will be in orange colours and the activations will be in black and white.
Brighten - brightens all the graphical output in the window.
Darken - darkens all graphical output in the window.
Cut:
After pressing this button (at the top of the graphics window), whatever image (or other object) you click on is removed.
Move:
After pressing this button (at the top of the graphics window), whatever image (or other object) you click on and hold down the left hand mouse button can be moved anywhere in the graphics window. Release the mouse button to drop the image to a specified location.
Size:
After pressing this button (at the top of the graphics window), click on an image or object with the left-hand mouse button to make it smaller, or click on it with the middle button to enlarge it.
Text:
Pressing "Text" at the top of the graphics window will change the cursor to a set square. Click on the left mouse button where you want the lower left of the text to start on the graphical page. Then type in the desired text to describe the graphical output.
Technical notes
Z scores and p values
p values have a direct and unchangeable relationship to the Z scores, as defined by the normal distribution. Thus a p value of 0.05 gives a Z score of 1.64, p=0.01 gives Z=2.33. and p=0.001 gives Z=3.09. In case you were interested, these values can be obtained from the matlab window thus:
>> spm_invNcdf(1 - [0.05 0.01 0.001] )
ans =
1.6449 2.3263 3.0902
Corrected extent thresholding
During the Results input section, you will be asked whether you want a corrected extent threshold:"Use corrected extent threshold"? We suggest that you routinely choose no to this question. If you chose yes, then the subsequent spm map will be thresholded for corrected extent. Let's say you type in a corrected p value here of 0.05. This means that spm will only allow to be displayed, clusters in the map that are large enough, that a cluster of this size would be predicted to occur by chance on only 5% of occasions.
More technically, spm estimates, using Random Field Theory, the size of cluster that is large enough that it would be expected to occur in only 5% of maps containing normally distributed random numbers with a similar spatial correlation to your spm map, and at the Z score height threshold that you specified earlier. It then removes all clusters smaller than the size of the cluster it has estimated.
This extent thresholding will severely limit the number of activation areas presented. Although this does give a useful conservative estimation of the significant signal in your image, we prefer the corrected height threshold for this purpose. You can use this by following the procedure below.
Corrected height thresholding
What if you want to threshold your spm map using the random field height correction? You can do this, by finding out the threshold before you press the Results button. If you are using SPM on the CBU network, you can type, in the matlab window:
>> corr_zthresh
(Users outside the CBU will need to download the files corr_zthresh.m and zspm_P.m to a directory on their matlab path in order to use this command).
The program will then ask you for the directory of the analysis files, and the corrected height p value that you want to threshold at. After you have entered this information, it will print out the relevant Z threshold in the matlab window. Now, when you enter the results section, type this Z value into the Height threshold box, and refuse any further thresholding in the subsequent inputs, and your Z map will be thresholded only for corrected height.
Maximum intensity projections (glass brain views)
The "glass brain views" of spm are technically known as Maximum Intensity Projections (MIPs). This is because the gray scale of the activation for each pixel of each view of the brain is proportional to the the largest value (be it Z score or F ratio), for the voxel with the maximun intensity, of all the voxels on a line going through the position of that pixel, perpendicular to the plane of the page. Thus a dark dot that is apparently in the lateral temporal cortex on the sagittal view might in fact represent a very activated voxel in the left, or right temporal cortices, or any bit of brain (or CSF!) in between.
Daniel Bor and Matthew Brett 11/5/99
