SPM5 includes a normalization method in the segmentation options that relies on segmenting the image and normalizating the segmented gray matter. In fact this is the standard AA normalisation routine (Automatic Analysis normalisation with segment).
This method may fail in certain cases. The example below illustrates it: at the bottom, the normalised brain on the left is smaller and has not been reoriented correctly.
If you look at the bottom right figure, supposedly of the GM segment, you will see that a lot of her scalp has been segmented to GM by error.
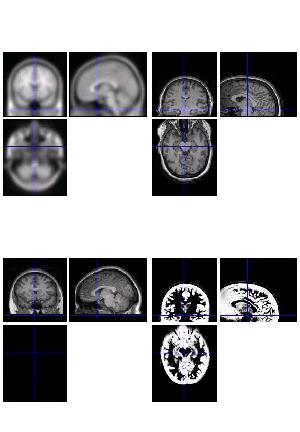
In order to allow monitoring of such occurrences, please mail to imagerstech@mrc-cbu.cam.ac.uk if you notice this problem.
The most likely problem in this specific case was (1) there was a lot of neck and shoulders, with some artifacts, near the bottom of the image (i.e, the field-of-view was probably too large) and (2) the image was a long way from the template (e.g, bad positioning of field-of-view).
The best solution, though involving manual intervention, is to reposition the structural (via Display) so that it is closer to the template (ie origin = AC). More details on how to do this can be found here: http://imaging.mrc-cbu.cam.ac.uk/meg/RepositioningMRIs. Note that any functionals coregistered with the structural will also need to be repositioned the same way (apply the reorientation parameters to the functional as well as structural images via Display).
If you want to stick with automated preprocessing, another solution is to use the SPM5 GUI 'normalize' procedure (i.e, the same normalisation as in SPM2) instead of the SPM5 'segment' procedure (ie use aamod_norm not aamod_norm_noss).